| Year |
Title |
Authors |
Journal |
Links |
| 2025 | Fundamental limits on patterning by Turing-like reaction-diffusion mechanisms. | Muzatko D, Daga B, Hiscock TW† | biorXiv | pdf |
| 2024 | A dynamic Hedgehog gradient orients tracheal cartilage rings. | Kingsley EP, Mishkind D, Hiscock TW†, Tabin CJ† | biorXiv | pdf |
| 2024 | Self-organized BMP signaling dynamics underlie the development and evolution of digit segmentation patterns in birds and mammals. | Grall E, Feregrino C, Fischer S, De Courten A, Sacher F, Hiscock TW†, Tschopp P† | PNAS | pdf |
| 2021 | Secreted inhibitors drive the loss of regeneration competence in Xenopus limbs. | Aztekin C*, Hiscock TW*, Gurdon J, Jullien J, Marioni J, Simons, BD | Development | pdf |
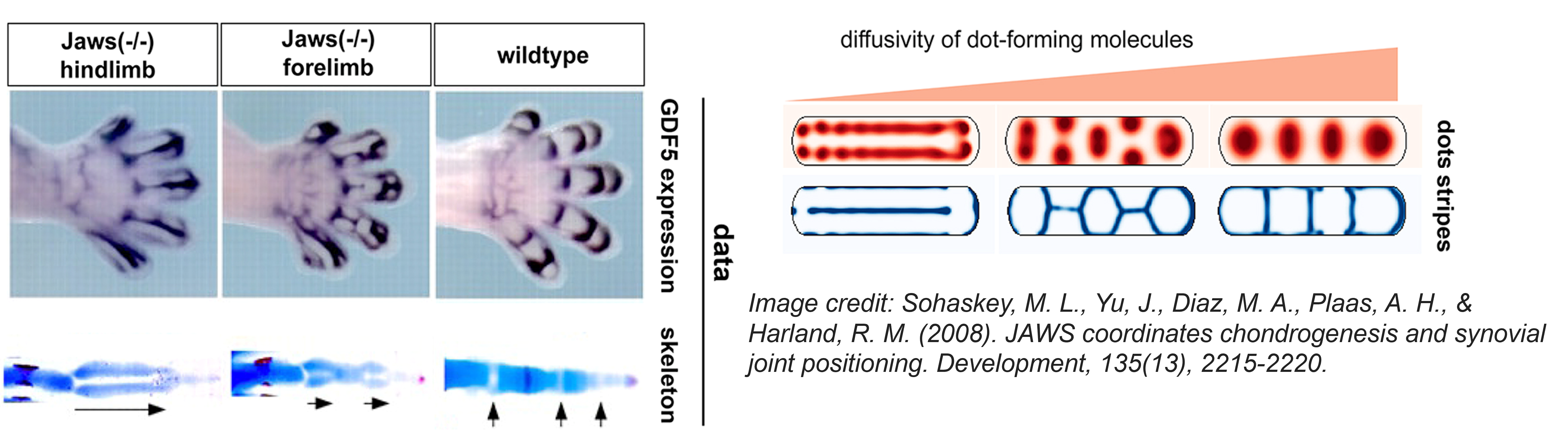
| 2020 | A dot-stripe model of joint patterning in the tetrapod limb. | Cornwall Scoones J, Hiscock TW† | Development | pdf |
| 2020 | The myeloid lineage is required for the emergence of a regeneration-permissive environment following Xenopus tail amputation. | Aztekin C, Hiscock TW, Butler R, Andino FDJ, Robert J, Gurdon JB, Jullien J. | Development | pdf |
| 2019 | Adapting machine-learning algorithms to design gene circuits. | Hiscock TW† | BMC bioinformatics. | pdf |
| 2019 | Navigating at night: fundamental limits on the sensitivity of radical pair magnetoreception under dim light. | Hiscock HG, Hiscock TW, Kattnig DR, Scrivener T, Lewis AM, Manolopoulos DE, Hore PJ. | Quarterly reviews of biophysics | pdf |
| 2019 | Identification of a regeneration-organizing cell in the Xenopus tail. | Aztekin C*, Hiscock TW*, Marioni JC, Gurdon JB, Simons BD, Jullien J. | Science | pdf |
| 2019 | A single-cell molecular map of mouse gastrulation and early organogenesis. | Pijuan-Sala B, Griffiths JA, Guibentif C, Hiscock TW, Jawaid W, Calero-Nieto FJ, et al. | Nature | link |
| 2018 | Feedback between tissue packing and neurogenesis in the zebrafish neural tube. | Hiscock TW, Miesfeld JB, Mosaliganti KR, Link BA, Megason SG. | Development | pdf |
| 2018 | Size-reduced embryos reveal a gradient scaling-based mechanism for zebrafish somite formation. | Ishimatsu K, Hiscock TW, Collins ZM, Sari DWK, Lischer K, Richmond DL, et al. | Development | pdf |
| 2018 | Identity and novelty in the avian syrinx. | Kingsley EP, Eliason CM, Riede T, Li Z, Hiscock TW, Farnsworth M, et al. | PNAS | pdf |
| 2018 | Observing the cell in its native state: Imaging subcellular dynamics in multicellular organisms. | Liu T, Upadhyayula S et al. | Science | pdf |
| 2017 | On the Formation of Digits and Joints during Limb Development. | Hiscock TW, Tschopp P, Tabin CJ. | Dev Cell | pdf |
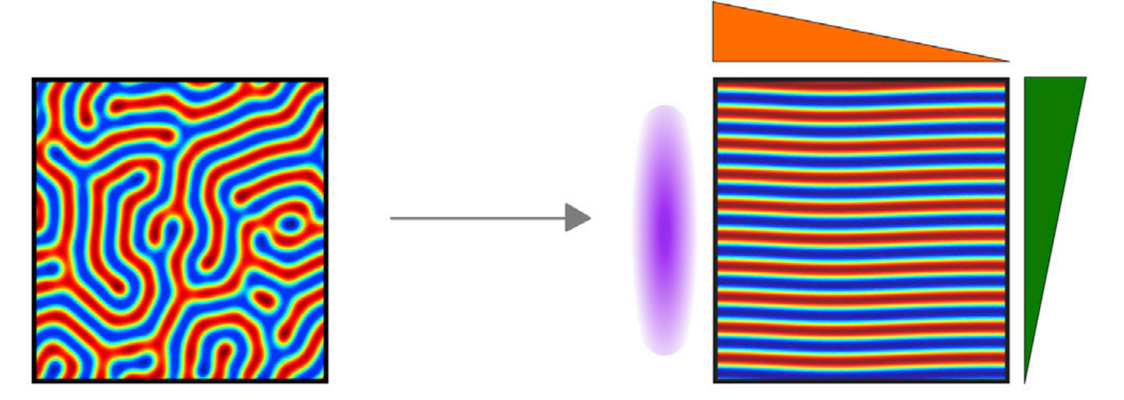
| 2015 | Orientation of Turing-like Patterns by Morphogen Gradients and Tissue Anisotropies. | Hiscock TW†, Megason SG† | Cell Systems | pdf |
| 2015 | Mathematically guided approaches to distinguish models of periodic patterning. | Hiscock TW, Megason SG | Development | pdf |
| 2014 | Interplay of cell shape and division orientation promotes robust morphogenesis of developing epithelia. | Xiong F, Ma W, Hiscock TW, Mosaliganti KR, Tentner AR, Brakke KA, et al. | Cell | pdf |